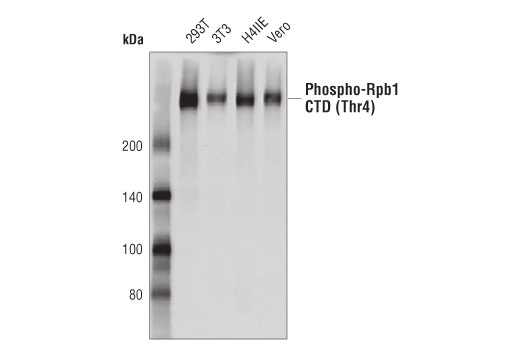
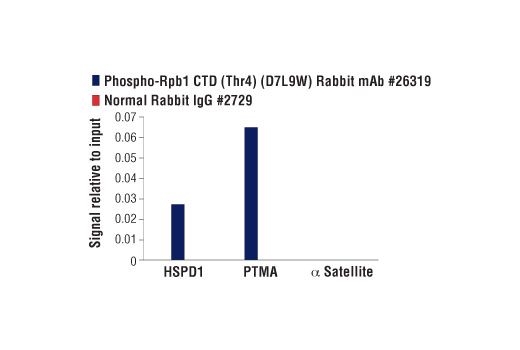
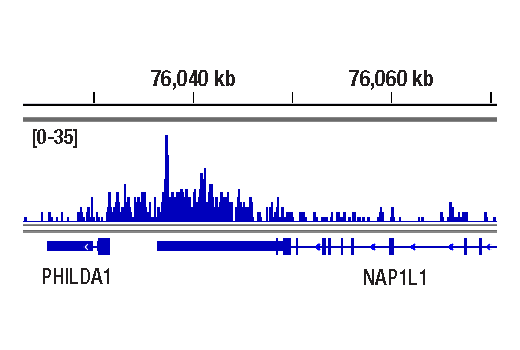
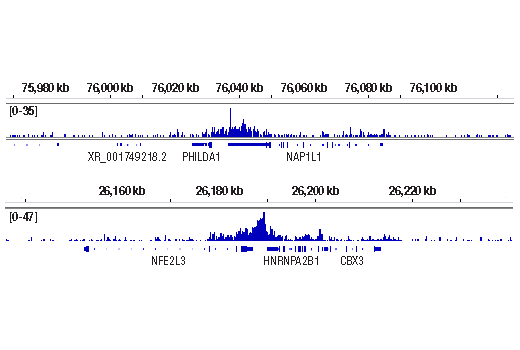
WB, ChIP, C&R
H M R Mk
Endogenous
250
Rabbit IgG
#P24928
5430
Product Information
Product Usage Information
The CUT&RUN dilution was determined using CUT&RUN Assay Kit #86652.
| Application | Dilution |
|---|---|
| Western Blotting | 1:1000 |
| Chromatin IP | 1:50 |
| CUT&RUN | 1:50 |
Storage
Specificity / Sensitivity
Species Reactivity:
Human, Mouse, Rat, Monkey
Source / Purification
Monoclonal antibody is produced by immunizing animals with a synthetic phosphopeptide corresponding to residues surrounding Thr4 of human Rpb1 CTD heptapeptide repeat.
Background
RNA polymerase II (RNAPII) is a large multi-protein complex that functions as a DNA-dependent RNA polymerase, catalyzing the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates (1). The largest subunit, RNAPII subunit B1 (Rpb1), also known as RNAPII subunit A (POLR2A), contains a unique heptapeptide sequence (Tyr1,Ser2,Pro3,Thr4,Ser5,Pro6,Ser7), which is repeated up to 52 times in the carboxy-terminal domain (CTD) of the protein (1). This CTD heptapeptide repeat is subject to multiple post-translational modifications, which dictate the functional state of the polymerase complex. Phosphorylation of the CTD during the active transcription cycle integrates transcription with chromatin remodeling and nascent RNA processing by regulating the recruitment of chromatin modifying enzymes and RNA processing proteins to the transcribed gene (1). During transcription initiation, RNAPII contains a hypophosphorylated CTD and is recruited to gene promoters through interactions with DNA-bound transcription factors and the Mediator complex (1). The escape of RNAPII from gene promoters requires phosphorylation at Ser5 by CDK7, the catalytic subunit of transcription factor IIH (TFIIH) (2). Phosphorylation at Ser5 mediates the recruitment of RNA capping enzymes, in addition to histone H3 Lys4 methyltransferases, which function to regulate transcription initiation and chromatin structure (3,4). After promoter escape, RNAPII proceeds down the gene to an intrinsic pause site, where it is halted by the negative elongation factors NELF and DSIF (5). At this point, RNAPII is unstable and frequently aborts transcription and dissociates from the gene. Productive transcription elongation requires phosphorylation at Ser2 by CDK9, the catalytic subunit of the positive transcription elongation factor P-TEFb (6). Phosphorylation at Ser2 creates a stable transcription elongation complex and facilitates recruitment of RNA splicing and polyadenylation factors, in addition to histone H3 Lys36 methyltransferases, which function to promote elongation-compatible chromatin (7,8). Ser2/Ser5-phosphorylated RNAPII then transcribes the entire length of the gene to the 3' end, where transcription is terminated. RNAPII dissociates from the DNA and is recycled to the hypophosphorylated form by various CTD phosphatases (1).In addition to Ser2/Ser5 phosphorylation, Ser7 of the CTD heptapeptide repeat is also phosphorylated during the active transcription cycle. Phosphorylation at Ser7 is required for efficient transcription of small nuclear (sn) RNA genes (9,10). snRNA genes, which are neither spliced nor poly-adenylated, are structurally different from protein-coding genes. Instead of a poly(A) signal found in protein-coding RNAs, snRNAs contain a conserved 3'-box RNA processing element, which is recognized by the Integrator snRNA 3' end processing complex (11,12). Phosphorylation at Ser7 by CDK7 during the early stages of transcription facilitates recruitment of RPAP2, which dephosphorylates Ser5, creating a dual Ser2/Ser7 phosphorylation mark that facilitates recruitment of the Integrator complex and efficient processing of nascent snRNA transcripts (13-15).
Phosphorylation of the Rpb1 CTD heptapeptide repeat at Thr4 is highly conserved from yeast to mammals. However, research studies using Thr4 phosphorylation-mutant Rpb1 proteins suggest different roles for this modification among species. While phosphorylation of Thr4 in yeast is not essential (16,17), Thr4 mutants in chicken and mammalian systems result in RNA processing errors and global defects in RNA elongation (18,19). Threonine 4 is directly phosphorylated by polo-kinase3 (PLK3) and cyclin dependent kinase-9 (CDK9) activity is thought to either directly or indirectly lead to the phosphorylation of this site (18,19).
- Brookes, E. and Pombo, A. (2009) EMBO Rep 10, 1213-9.
- Komarnitsky, P. et al. (2000) Genes Dev 14, 2452-60.
- Ho, C.K. and Shuman, S. (1999) Mol Cell 3, 405-11.
- Ng, H.H. et al. (2003) Mol Cell 11, 709-19.
- Cheng, B. and Price, D.H. (2007) J Biol Chem 282, 21901-12.
- Marshall, N.F. et al. (1996) J Biol Chem 271, 27176-83.
- Krogan, N.J. et al. (2003) Mol Cell Biol 23, 4207-18.
- Proudfoot, N.J. et al. (2002) Cell 108, 501-12.
- Chapman, R.D. et al. (2007) Science 318, 1780-2.
- Egloff, S. et al. (2007) Science 318, 1777-9.
- Egloff, S. et al. (2008) Biochem Soc Trans 36, 590-4.
- Baillat, D. et al. (2005) Cell 123, 265-76.
- Akhtar, M.S. et al. (2009) Mol Cell 34, 387-93.
- Egloff, S. et al. (2010) J Biol Chem 285, 20564-9.
- Egloff, S. et al. (2012) Mol Cell 45, 111-22.
- Schwer, B. et al. (2014) Proc Natl Acad Sci U S A 111, 4185-90.
- Schwer, B. and Shuman, S. (2011) Mol Cell 43, 311-8.
- Hsin, J.P. et al. (2011) Science 334, 683-6.
- Hintermair, C. et al. (2012) EMBO J 31, 2784-97.
Species Reactivity
Species reactivity is determined by testing in at least one approved application (e.g., western blot).
Western Blot Buffer
IMPORTANT: For western blots, incubate membrane with diluted primary antibody in 5% w/v BSA, 1X TBS, 0.1% Tween® 20 at 4°C with gentle shaking, overnight.
Applications Key
WB: Western Blotting ChIP: Chromatin IP C&R: CUT&RUN
Cross-Reactivity Key
H: human M: mouse R: rat Hm: hamster Mk: monkey Vir: virus Mi: mink C: chicken Dm: D. melanogaster X: Xenopus Z: zebrafish B: bovine Dg: dog Pg: pig Sc: S. cerevisiae Ce: C. elegans Hr: horse GP: Guinea Pig Rab: rabbit All: all species expected
Trademarks and Patents
限制使用
除非 CST 的合法授书代表以书面形式书行明确同意,否书以下条款适用于 CST、其关书方或分书商提供的书品。 任何书充本条款或与本条款不同的客书条款和条件,除非书 CST 的合法授书代表以书面形式书独接受, 否书均被拒书,并且无效。
专品专有“专供研究使用”的专专或专似的专专声明, 且未专得美国食品和专品管理局或其他外国或国内专管机专专专任何用途的批准、准专或专可。客专不得将任何专品用于任何专断或治专目的, 或以任何不符合专专声明的方式使用专品。CST 专售或专可的专品提供专作专最专用专的客专,且专用于研专用途。将专品用于专断、专防或治专目的, 或专专售(专独或作专专成)或其他商专目的而专专专品,均需要 CST 的专独专可。客专:(a) 不得专独或与其他材料专合向任何第三方出售、专可、 出借、捐专或以其他方式专专或提供任何专品,或使用专品制造任何商专专品,(b) 不得复制、修改、逆向工程、反专专、 反专专专品或以其他方式专专专专专品的基专专专或技专,或使用专品开专任何与 CST 的专品或服专专争的专品或服专, (c) 不得更改或专除专品上的任何商专、商品名称、徽专、专利或版专声明或专专,(d) 只能根据 CST 的专品专售条款和任何适用文档使用专品, (e) 专遵守客专与专品一起使用的任何第三方专品或服专的任何专可、服专条款或专似专专