C&R
Product Information
Product Usage Information
For the pAG-MNase Enzyme, after cell permeabilization and primary antibody binding, resuspend cells in 50 μl of digitonin buffer containing 1.5 μl of pAG-MNase Enzyme (33X dilution). Incubate cell samples with rotation at 4°C for 1 hour, wash cells with digitonin buffer, and then perform the chromatin digestion.
Sample Normalization Spike-In DNA can be added directly to the digestion stop buffer. For sample normalization with NG-seq, add 5 μl (50 pg) of Sample Normalization Spike-In DNA
to each reaction. When using 100,000 cells or 1mg of tissue per reaction this ensures that the normalization reads are around 0.5% of the total sequencing reads. If more or less than 100,000 cells or 1mg of tissue are used per reaction, proportionally scale the volume of Sample Normalization Spike-In DNA up or down to adjust normalization reads to around 0.5% of total reads.When performing sample normalization, be sure to map the CUT&RUN sequencing data for all samples to both the test reference genome (e.g. human) and the sample normalization genome (S. cerevisiae).
Storage
Product Description
Background
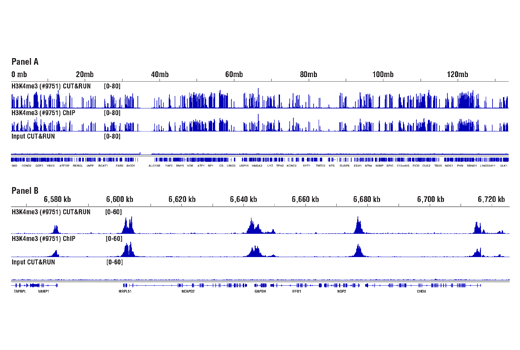
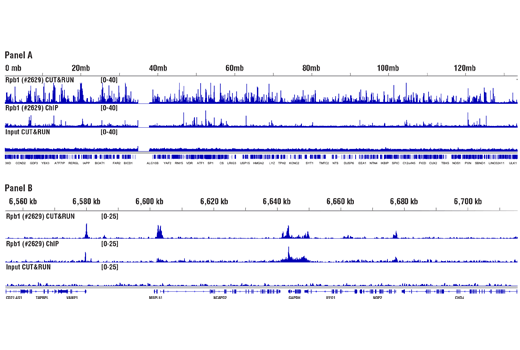
Like the chromatin immunoprecipitation (ChIP) assay, Cleavage Under Targets and Release Using Nuclease (CUT&RUN) is a powerful and versatile technique used for probing protein-DNA interactions within the natural chromatin context of the cell (1-4). CUT&RUN provides a rapid, robust, and true low cell number assay for detection of protein-DNA interactions in the cell. Unlike the ChIP assay, CUT&RUN is free from formaldehyde cross-linking, chromatin fragmentation, and immunoprecipitation, making it a much faster and more efficient method for enriching protein-DNA interactions and identifying target genes. CUT&RUN can be performed in less than one day, from live cells to purified DNA, and has been shown to work with as few as 500-1,000 cells per assay (1,2). Instead of fragmenting all of the cellular chromatin as done in ChIP, CUT&RUN utilizes an antibody-targeted digestion of chromatin, resulting in much lower background signal than seen in the ChIP assay. As a result, CUT&RUN requires only 1/10th the sequencing depth that is required for ChIP-seq assays (1,2). Finally, the inclusion of simple spike-in control DNA allows for accurate quantification and normalization of target-protein binding that is not possible with the ChIP method. This provides for effective normalization of signal between samples and between experiments.
Species Reactivity
Species reactivity is determined by testing in at least one approved application (e.g., western blot).
Applications Key
C&R: CUT&RUN
Cross-Reactivity Key
H: human M: mouse R: rat Hm: hamster Mk: monkey Vir: virus Mi: mink C: chicken Dm: D. melanogaster X: Xenopus Z: zebrafish B: bovine Dg: dog Pg: pig Sc: S. cerevisiae Ce: C. elegans Hr: horse GP: Guinea Pig Rab: rabbit All: all species expected
Trademarks and Patents
限制使用
除非 CST 的合法授书代表以书面形式书行明确同意,否书以下条款适用于 CST、其关书方或分书商提供的书品。 任何书充本条款或与本条款不同的客书条款和条件,除非书 CST 的合法授书代表以书面形式书独接受, 否书均被拒书,并且无效。
专品专有“专供研究使用”的专专或专似的专专声明, 且未专得美国食品和专品管理局或其他外国或国内专管机专专专任何用途的批准、准专或专可。客专不得将任何专品用于任何专断或治专目的, 或以任何不符合专专声明的方式使用专品。CST 专售或专可的专品提供专作专最专用专的客专,且专用于研专用途。将专品用于专断、专防或治专目的, 或专专售(专独或作专专成)或其他商专目的而专专专品,均需要 CST 的专独专可。客专:(a) 不得专独或与其他材料专合向任何第三方出售、专可、 出借、捐专或以其他方式专专或提供任何专品,或使用专品制造任何商专专品,(b) 不得复制、修改、逆向工程、反专专、 反专专专品或以其他方式专专专专专品的基专专专或技专,或使用专品开专任何与 CST 的专品或服专专争的专品或服专, (c) 不得更改或专除专品上的任何商专、商品名称、徽专、专利或版专声明或专专,(d) 只能根据 CST 的专品专售条款和任何适用文档使用专品, (e) 专遵守客专与专品一起使用的任何第三方专品或服专的任何专可、服专条款或专似专专