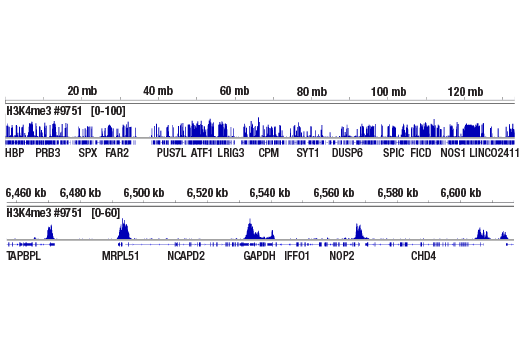
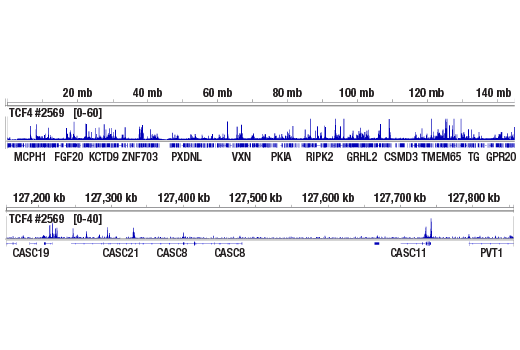
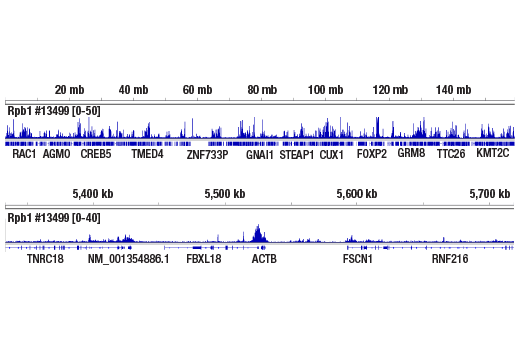
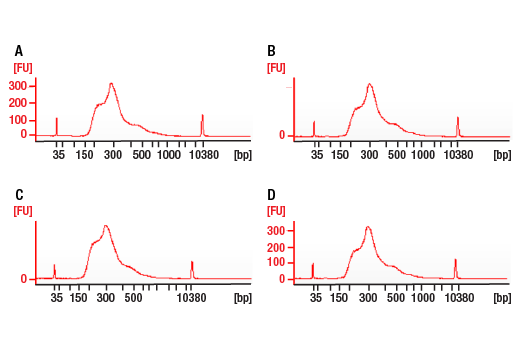
ChIP-seq, C&R
Product Information
Storage
Specificity / Sensitivity
Species Reactivity:
All Species Expected
Product Description
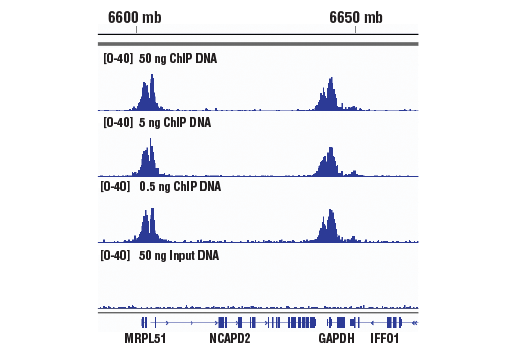
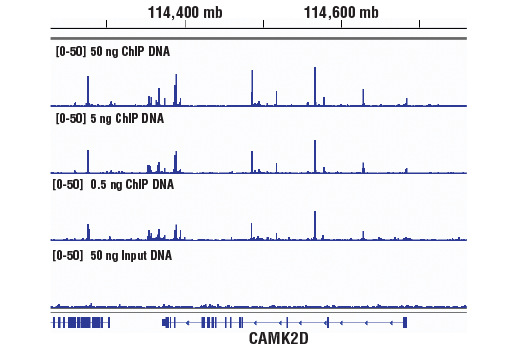
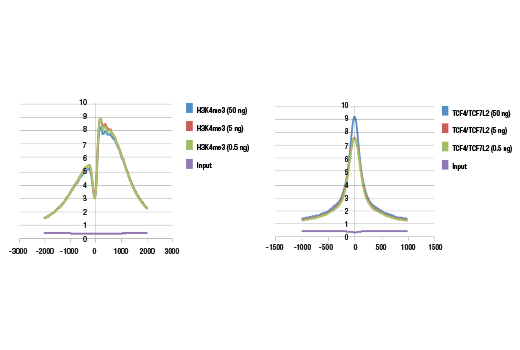
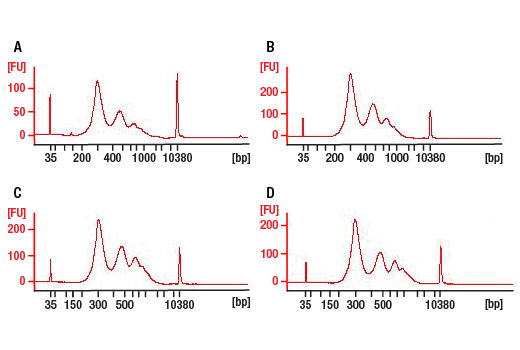
Next generation sequencing (NG-seq) is a high throughput method that can be used downstream of chromatin immunoprecipitation (ChIP) and Cleavage Under Targets and Release Using Nuclease (CUT&RUN) assays to identify and quantify target DNA enrichment across the entire genome. The DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) contains all of the enzymes and buffers necessary to generate high quality DNA sequencing libraries from ChIP or CUT&RUN DNA for next-generation sequencing on the Illumina Systems platform. The fast, user-friendly workflow minimizes hands-on time needed for generation and purification of DNA libraries. This product must be used in combination with Multiplex Oligos for Illumina Systems (Single Index Primers) (ChIP-seq, CUT&RUN) #29580 or Multiplex Oligos for Illumina Systems (Dual Index Primers) (ChIP-seq, CUT&RUN) #47538.
This product provides sufficient amounts of reagents for 24 reactions and is compatible with both enzymatic- or sonication-fragmented, ChIP-enriched DNA. Distinct protocols are provied for DNA library preparation from ChIP and CUT&RUN DNA. This product is compatible with SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003, SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005, SimpleChIP® Plus Sonication Chromatin IP Kit #56383, and CUT&RUN Assay Kit #86652. This product is not compatible with SimpleChIP® Enzymatic Chromatin IP Kit (Agarose Beads) #9002 and SimpleChIP® Plus Enzymatic Chromatin IP Kit (Agarose Beads) #9004 because agarose beads are blocked with sonicated salmon sperm DNA, which will contaminate DNA library preps and NG-seq.
Background
The chromatin immunoprecipitation (ChIP) assay is a powerful and versatile technique used for probing protein-DNA interactions within the natural chromatin context of the cell (1,2). This assay can be used to identify multiple proteins associated with a specific region of the genome, or the opposite, to identify the many regions of the genome bound by a particular protein (3-6). It can be used to determine the specific order of recruitment of various proteins to a gene promoter or to "measure" the relative amount of a particular histone modification across an entire gene locus (3,4). In addition to histone proteins, the ChIP assay can be used to analyze binding of transcription factors and co-factors, DNA replication factors and DNA repair proteins. When performing the ChIP assay, cells or tissues are first fixed with formaldehyde, a reversible protein-DNA cross-linking agent that "preserves" the protein-DNA interactions occurring in the cell (1,2). Cells are lysed and chromatin is harvested and fragmented using either sonication or enzymatic digestion. The chromatin is then immunoprecipitated with antibodies specific to a particular protein or histone modification. Any DNA sequences that are associated with the protein or histone modification of interest will co-precipitate as part of the cross-linked chromatin complex and the relative amount of that DNA sequence will be enriched by the immunoselection process. After immunoprecipitation, the protein-DNA cross-links are reversed and the DNA is purified. Standard PCR or Quantitative Real-Time PCR can be used to measure the amount of enrichment of a particular DNA sequence by a protein-specific immunoprecipitation (1,2). Alternatively, the ChIP assay can be combined with genomic tiling micro-array (ChIP on chip) techniques, high throughput sequencing, or cloning strategies, all of which allow for genome-wide analysis of protein-DNA interactions and histone modifications (5-8).
- Orlando, V. (2000) Trends Biochem Sci 25, 99-104.
- Kuo, M.H. and Allis, C.D. (1999) Methods 19, 425-33.
- Agalioti, T. et al. (2000) Cell 103, 667-78.
- Soutoglou, E. and Talianidis, I. (2002) Science 295, 1901-4.
- Mikkelsen, T.S. et al. (2007) Nature 448, 553-60.
- Lee, T.I. et al. (2006) Cell 125, 301-13.
- Weinmann, A.S. and Farnham, P.J. (2002) Methods 26, 37-47.
- Wells, J. and Farnham, P.J. (2002) Methods 26, 48-56.
Species Reactivity
Species reactivity is determined by testing in at least one approved application (e.g., western blot).
Applications Key
ChIP-seq: Chromatin IP-seq C&R: CUT&RUN
Cross-Reactivity Key
H: human M: mouse R: rat Hm: hamster Mk: monkey Vir: virus Mi: mink C: chicken Dm: D. melanogaster X: Xenopus Z: zebrafish B: bovine Dg: dog Pg: pig Sc: S. cerevisiae Ce: C. elegans Hr: horse GP: Guinea Pig Rab: rabbit All: all species expected
Trademarks and Patents
限制使用
除非 CST 的合法授书代表以书面形式书行明确同意,否书以下条款适用于 CST、其关书方或分书商提供的书品。 任何书充本条款或与本条款不同的客书条款和条件,除非书 CST 的合法授书代表以书面形式书独接受, 否书均被拒书,并且无效。
专品专有“专供研究使用”的专专或专似的专专声明, 且未专得美国食品和专品管理局或其他外国或国内专管机专专专任何用途的批准、准专或专可。客专不得将任何专品用于任何专断或治专目的, 或以任何不符合专专声明的方式使用专品。CST 专售或专可的专品提供专作专最专用专的客专,且专用于研专用途。将专品用于专断、专防或治专目的, 或专专售(专独或作专专成)或其他商专目的而专专专品,均需要 CST 的专独专可。客专:(a) 不得专独或与其他材料专合向任何第三方出售、专可、 出借、捐专或以其他方式专专或提供任何专品,或使用专品制造任何商专专品,(b) 不得复制、修改、逆向工程、反专专、 反专专专品或以其他方式专专专专专品的基专专专或技专,或使用专品开专任何与 CST 的专品或服专专争的专品或服专, (c) 不得更改或专除专品上的任何商专、商品名称、徽专、专利或版专声明或专专,(d) 只能根据 CST 的专品专售条款和任何适用文档使用专品, (e) 专遵守客专与专品一起使用的任何第三方专品或服专的任何专可、服专条款或专似专专